
The “Jmol Application-Beginner” workshop will be 5-hours of watching relevant pre-recorded Spoken Tutorial videos with side-by-side learning, hands-on training, and interaction with experts.
#Install jmol registration
The registration form will be available until 18 September 2020, 2.00 PM. The interested participants have to register online and pay the registration fee. Useful for students, educators, and researchers in chemistry, biochemistry and pharmacy.
It is useful for classroom teaching and presentations. It is compatible with Windows, Mac and Linux Operating Systems.
#Install jmol software
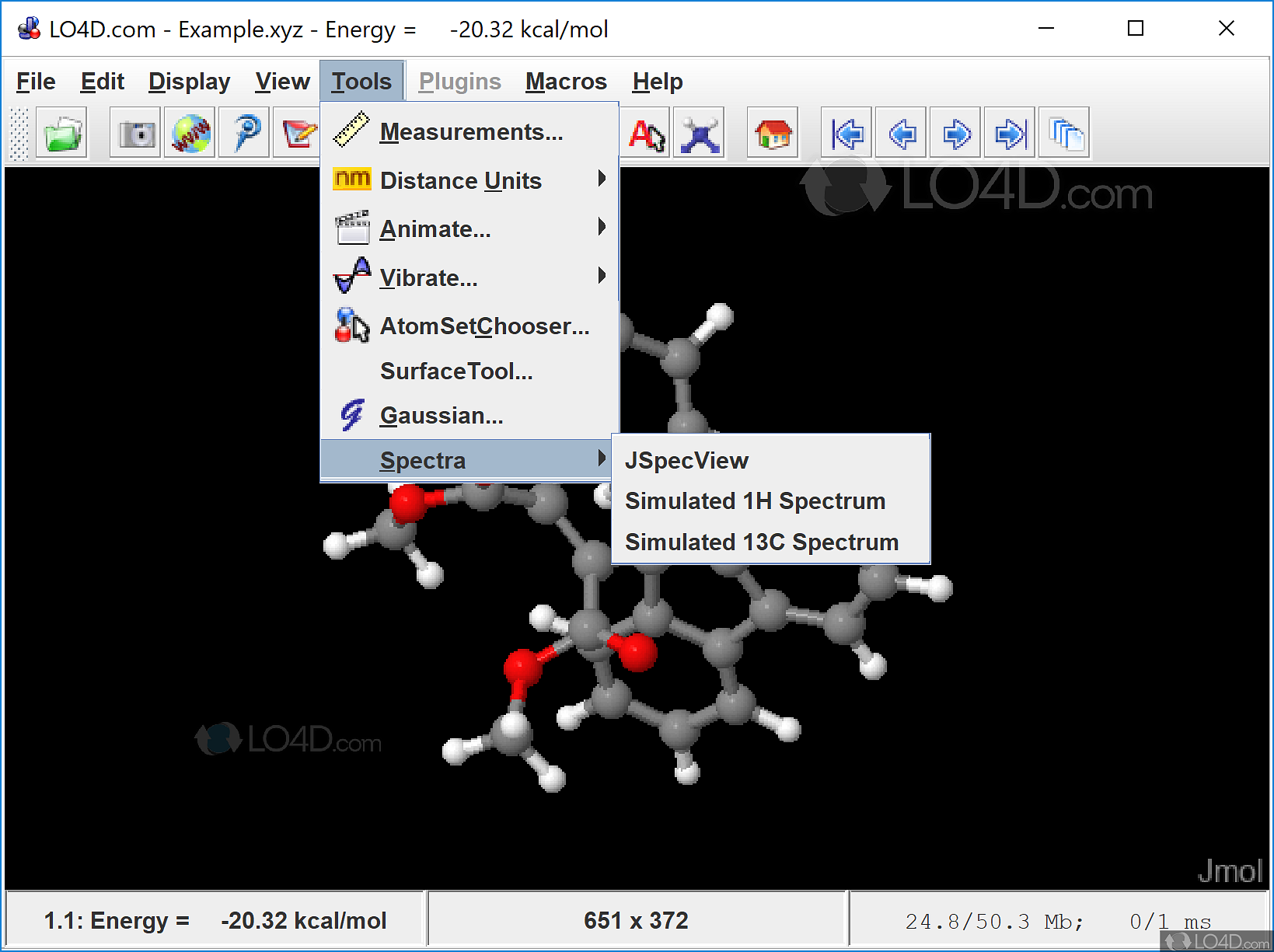
Jmol Application is a free and open source software developed in Java to view, create, and edit three-dimensional models of chemical structures, macromolecules (Crystal structures, Proteins, and Nucleic acids). Then you need to edit /artefact/file/lib.php to set the appropriate file mime-types.The international participants are requested to read the instructions and payment guidelines before filling the Google form for Jmol workshop registration $string = 'XYZ Coordinate Animation Format' $string = 'MacroMolecular Crystallographic Information File' $string = 'Crystallographic Information File' artefact/file/lang/en.utf8/Īdd, immediately after the other file types pdb.gz) files then you need to edit /lib/web.php to comment out the line that loads jQueryĪnd load JSmoljQuery instead. If you wish to display binary format (.pse or.
#Install jmol download
Some other edits of Mahara files are required (or you can copy across the appropriate files from the download bundle).
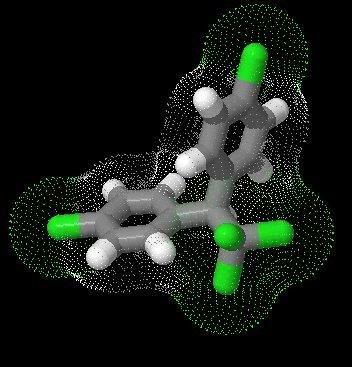
On unpacking, the /jmol folder (and its contents) is copied into the /artefact/file/blocktype folder of your Mahara installation. The latest version of the plugin, for Mahara 1.2, 1.3, 1.4, 1.5, 1.6 and 1.7 can be downloaded from Github NB Development has focused on functionality, and there may be security implications in running these scripts. Enter optional Jmol.js JavaScript commands to add custom controls below the applet ĭefault script values are applied for general use. Specify an optional Jmol startup script to customise initial display of the applet Ģ. Then upload/select the appropriate file and set the appropriate applet width and height (default 300 px v 300 px)įor more advanced Jmol use, there are a couple of textarea controls that allow you to:ġ. When editing the contents of a Mahara Page (View), drag the Jmol icon from the 'Files, images and videos' tab into your Page (View) Xyz chemical/x-xyz XYZ Coordinate Animation format Sdf chemical/x-mdl-sdfile Structure Data File Mol2 chemical/x-mol2 SYBYL Tripos Mol2 file format Mcif chemical/x-mmcif MacroMolecular Crystallographic Information File This plugin currently uses the following subset of chemical structure file (chemical MIME) types, though others can be easily added (see below)Ĭif chemical/x-cif Crystallographic Information FileĬml chemical/x-cml Chemical Markup Language Jmol/JSmol can use a variety of chemical structure file formats (chemical MIME types). It is similar to other Mahara artefact file blocktype plugins such as the Embedded media plugin. JSmol offers wider platform support than the Jmol Java applet. JSmol is the recently developed JavaScript/HTML5 version of Jmol. This plugin provides the interactive 3D display of chemical structure files, uploaded to Mahara, using Jmol/JSmol.

Mahara artefact file blocktype jmol/jsmol plugin


 0 kommentar(er)
0 kommentar(er)
